Best Way To Analyze Qpcr Data
The Applied Biosystems PresenceAbsence Analysis Module is part of a fast powerful flexible online data analysis solution that provides an online tool kit for analysis of qPCR data. A good assay should reveal a single peak in the plot depicting the negative first derivative of fluorescence over temperature -dFdT while additional peaks usually indicate the presence of non-specific amplification products.
 The Ultimate Qpcr Experiment Producing Publication Quality Reproducible Data The First Time Trends In Biotechnology
The Ultimate Qpcr Experiment Producing Publication Quality Reproducible Data The First Time Trends In Biotechnology
Monitor the qPCR reaction and a melt step is included after the amplification process in the qPCR program.

Best way to analyze qpcr data. The numbers it spits out are by the very nature of the process inverse log2 values. Double delta Ct analysis and the relative standard curve method Pfaffl method. It requires an experiment similar to that of calculating the standard curve.
This can be done by using a variety of software which can determine the best reference gene to use such as geNorm available in Biogazelles qbase program or Normfinder a free Excel add-on. There are two approaches to analyzing qPCR data. Serial dilutions of the genes of interest and controls are used as input to the reaction and different calculations are made.
01092014 The choice of the correct method to analyze the qPCR data at hand strongly depends on the experimental setup and assay. Positive Control IP Ct 27746 and Input Ct23956 Negative Control IP Ct304935 and. 11032021 There are two main ways to analyze qPCR data.
Data screening is the most important technique to check the nature of the data. Absolute quantification and relative quantification. 07082018 One way is to select the single best gene from the numerous ones tested to be used as the reference.
Absolute quantification allows the determination of how much DNA is in a given quantity of sample without performing comparative analyses with other samples. Therefore a number of review articles were recently published 15 72 to facilitate the choice of the most suitable analysis method for. I do not sure if it feets exactly your.
02042015 Ruijter has developed the program LinReg PCR freeware at HFRC that allows one of the best ways to analyse expression data. Ad Rapidly Characterize Molecular Target w Cancer Tissue qPCR Array. I am putting my positive and negative control Ct values.
One of the methods to screen the data is the Box-Cox Transformation. For the example analysis I will use the data below. Learn more about Applied Biosystems Analysis Modules Purchase the Applied Biosystems Analysis Modules after registering for a Thermo Fisher Cloud account.
26092017 How To Analyse ChIP qPCR Data There are a few ways in which you can analyse chromatin immunoprecipitation ChIP data acquired from quantitative real-time polymerase chain reaction qPCR. Httpwwwgene-quantificationinfo then go to Data Analysis qPCR software applications REST versions then scroll down to New REST software application are available Relative N E x CTxcontrol-sample E R CTRcontrol-sample E R CTsample E X CTsample E R calibrator E X calibrator. I just want to ask how to analyse ChIP PCR data.
Both methods make assumptions and have their limitations so the method you should use for your analysis will depend on your experimental design. Used in qPCR data analysis and a practical example using Integromics RealTime StatMiner the unique software analysis package specialized for qPCR experiments which is compatible with all Applied Biosystems Instruments. 01042020 Fortunately regardless of the method used in the analysis of qPCR data The quality assessment are done in a similar way.
Ad Rapidly Characterize Molecular Target w Cancer Tissue qPCR Array. This means 1 High numbers low. For statistical analysis of RT-qPCR we use a Bootstratio approach I included the links to the publication and the web where you could do the calculations.
Two of the most common ways to report ChIP qPCR are. QPCR uses an exponential process to generate meaningful quantitative data. Percentage of input and fold enrichment.
 Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
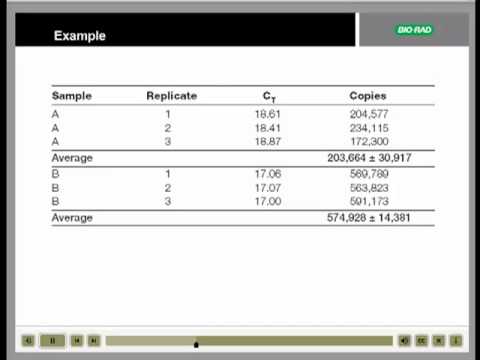 Real Time Qpcr Data Analysis Tutorial Youtube
Real Time Qpcr Data Analysis Tutorial Youtube
 Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
 Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
 Global Digital Pcr Dpcr And Real Time Pcr Qpcr Market 2018 By Manufacturers Countries Type And Application Forecast To 2023 Market Research Marketing Global
Global Digital Pcr Dpcr And Real Time Pcr Qpcr Market 2018 By Manufacturers Countries Type And Application Forecast To 2023 Market Research Marketing Global
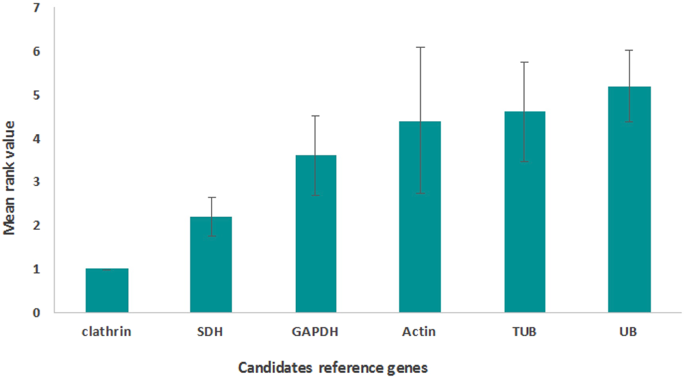 Identification Of Reference Genes For Rt Qpcr Data Normalization In Gammarus Fossarum Crustacea Amphipoda Scientific Reports X Mol
Identification Of Reference Genes For Rt Qpcr Data Normalization In Gammarus Fossarum Crustacea Amphipoda Scientific Reports X Mol
 To Identify Novel Splicing Variants From Exome Sequencing Data Researchers At University Of Milano Bicocca Italy Developed A Bio Point Mutation Data Mutation
To Identify Novel Splicing Variants From Exome Sequencing Data Researchers At University Of Milano Bicocca Italy Developed A Bio Point Mutation Data Mutation
 Auto Qpcr A Python Based Web App For Automated And Reproducible Analysis Of Qpcr Data Biorxiv
Auto Qpcr A Python Based Web App For Automated And Reproducible Analysis Of Qpcr Data Biorxiv

Plos One Stability Evaluation Of Reference Genes For Gene Expression Analysis By Rt Qpcr In Soybean Under Different Conditions
 Auto Qpcr A Python Based Web App For Automated And Reproducible Analysis Of Qpcr Data Biorxiv
Auto Qpcr A Python Based Web App For Automated And Reproducible Analysis Of Qpcr Data Biorxiv
 A Vic Hex Dye Alternative Ultra Bright And License Free Gene Expression Dye This Or That Questions
A Vic Hex Dye Alternative Ultra Bright And License Free Gene Expression Dye This Or That Questions
Https Www Qbaseplus Com Sites Default Files Publication Hellemans Vandesompele 2011 Pdf
 Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
Qpcr Data Analysis Better Results Through Iconoclasm Sciencedirect
 6 Quantitative Pcr The Deltadeltact Method Method The Real World Real
6 Quantitative Pcr The Deltadeltact Method Method The Real World Real
An Overview Of Technical Considerations When Using Quantitative Real Time Pcr Analysis Of Gene Expression In Human Exercise Research
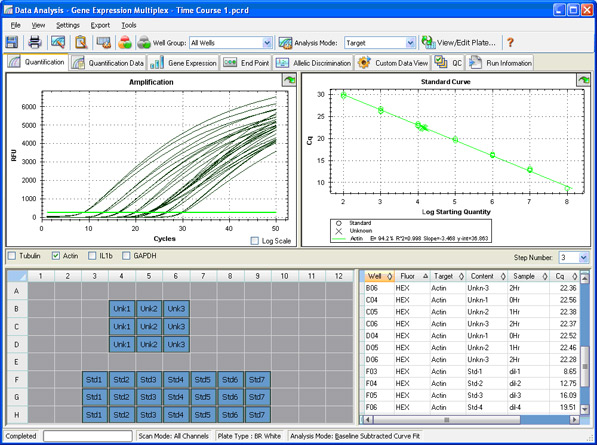 Introduction To Qpcr Instrumentation Lsr Bio Rad
Introduction To Qpcr Instrumentation Lsr Bio Rad
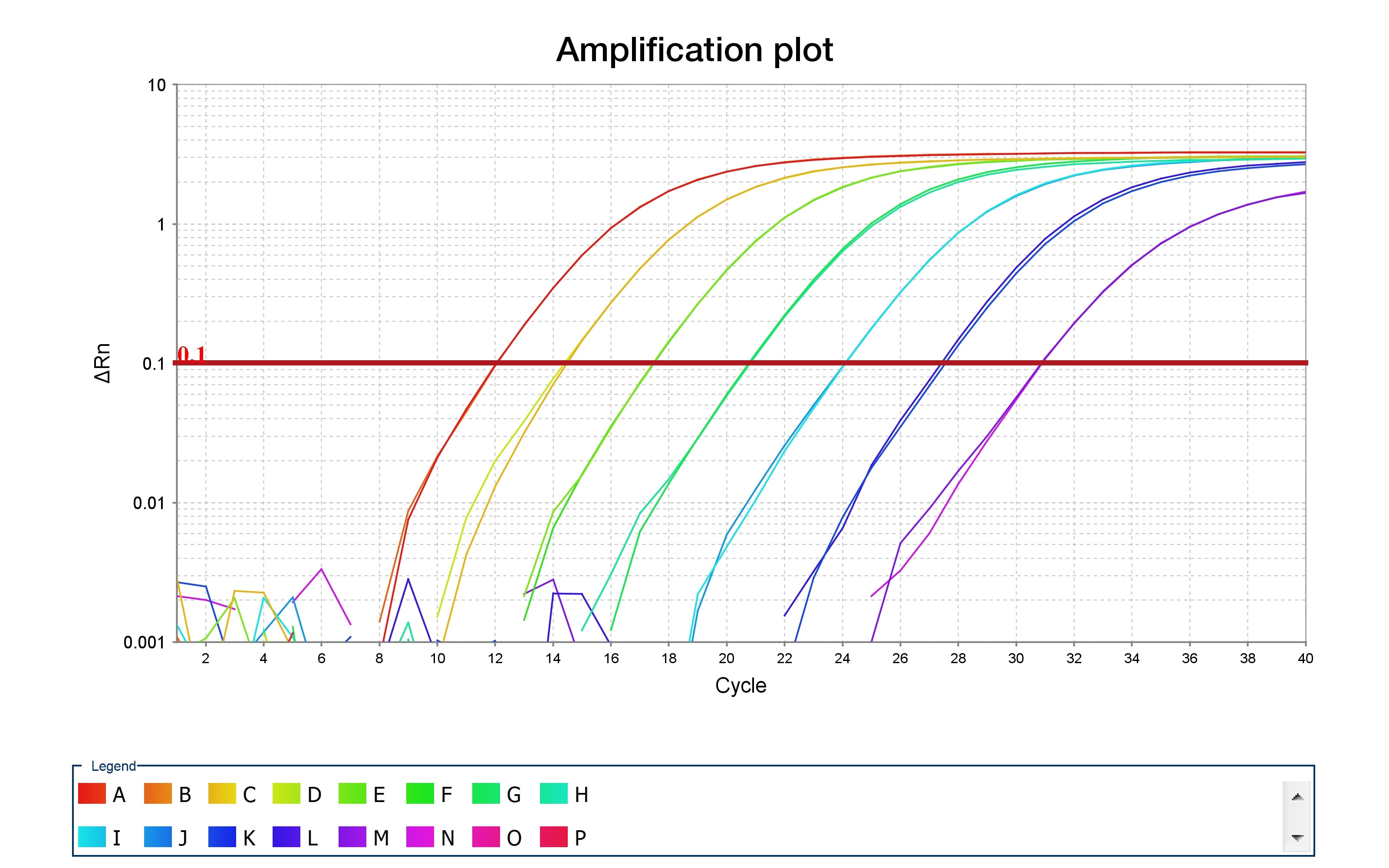 Ten Tips For Successful Qpcr Behind The Bench
Ten Tips For Successful Qpcr Behind The Bench
 4 Easy Steps To Analyze Your Qpcr Data Using Double Delta Ct Analysis Bitesize Bio Analysis Analyze Data
4 Easy Steps To Analyze Your Qpcr Data Using Double Delta Ct Analysis Bitesize Bio Analysis Analyze Data
0 Response to "Best Way To Analyze Qpcr Data"
Post a Comment